pertpy.tools.SCGEN¶
- class pertpy.tools.SCGEN(adata, n_hidden=800, n_latent=100, n_layers=2, dropout_rate=0.2, **model_kwargs)[source]¶
Jax Implementation of scGen model for batch removal and perturbation prediction.
Attributes table¶
Data attached to model instance. |
|
Manager instance associated with self.adata. |
|
The current device that the module's params are on. |
|
Returns computed metrics during training. |
|
Whether the model has been trained. |
|
Summary string of the model. |
|
Observations that are in test set. |
|
Observations that are in train set. |
|
Observations that are in validation set. |
Methods table¶
|
Removes batch effects. |
|
Converts a legacy saved model (<v0.15.0) to the updated save format. |
|
Deregisters the |
|
Retrieves the |
|
Get decoded expression. |
|
Returns the object in AnnData associated with the key in the data registry. |
|
Return the latent representation for each cell. |
|
Instantiate a model from the saved output. |
|
Return the full registry saved with the model. |
|
Predicts the cell type provided by the user in stimulated condition. |
|
Registers an |
|
Save the state of the model. |
|
Sets up the |
|
Move model to device. |
|
Train the model. |
|
Print summary of the setup for the initial AnnData or a given AnnData object. |
|
Print args used to setup a saved model. |
|
Plots the dot product between delta and latent representation of a linear classifier. |
|
Plots mean matching for a set of specified genes. |
|
Plots variance matching for a set of specified genes. |
Attributes¶
adata¶
- SCGEN.adata¶
Data attached to model instance.
adata_manager¶
- SCGEN.adata_manager¶
Manager instance associated with self.adata.
device¶
- SCGEN.device¶
history¶
- SCGEN.history¶
Returns computed metrics during training.
is_trained¶
- SCGEN.is_trained¶
Whether the model has been trained.
summary_string¶
- SCGEN.summary_string¶
Summary string of the model.
test_indices¶
- SCGEN.test_indices¶
Observations that are in test set.
train_indices¶
- SCGEN.train_indices¶
Observations that are in train set.
validation_indices¶
- SCGEN.validation_indices¶
Observations that are in validation set.
Methods¶
batch_removal¶
- SCGEN.batch_removal(adata=None)[source]¶
Removes batch effects.
- Parameters:
adata (
AnnData|None) – AnnData object with equivalent structure to initial AnnData. If None, defaults to the AnnData object used to initialize the model. Must have been setup with batch_key and labels_key, corresponding to batch and cell type metadata, respectively.- Returns:
~anndata.AnnData AnnData of corrected gene expression in adata.X and corrected latent space in adata.obsm[“latent”]. A reference to the original AnnData is in corrected.raw if the input adata had no raw attribute.
- Return type:
corrected
Examples
>>> import pertpy as pt >>> data = pt.dt.kang_2018() >>> pt.tl.SCGEN.setup_anndata(data, batch_key="label", labels_key="cell_type") >>> model = pt.tl.SCGEN(data) >>> model.train(max_epochs=10, batch_size=64, early_stopping=True, early_stopping_patience=5) >>> corrected_adata = model.batch_removal()
convert_legacy_save¶
- classmethod SCGEN.convert_legacy_save(dir_path, output_dir_path, overwrite=False, prefix=None, **save_kwargs)¶
Converts a legacy saved model (<v0.15.0) to the updated save format.
- Parameters:
dir_path (
str) – Path to directory where legacy model is saved.output_dir_path (
str) – Path to save converted save files.overwrite (
bool) – Overwrite existing data or not. IfFalseand directory already exists atoutput_dir_path, error will be raised.**save_kwargs – Keyword arguments passed into
save().
- Return type:
deregister_manager¶
- SCGEN.deregister_manager(adata=None)¶
Deregisters the
AnnDataManagerinstance associated with adata.If adata is None, deregisters all
AnnDataManagerinstances in both the class and instance-specific manager stores, except for the one associated with this model instance.
get_anndata_manager¶
- SCGEN.get_anndata_manager(adata, required=False)¶
Retrieves the
AnnDataManagerfor a given AnnData object specific to this model instance.Requires
self.idhas been set. Checks for anAnnDataManagerspecific to this model instance.
get_decoded_expression¶
- SCGEN.get_decoded_expression(adata=None, indices=None, batch_size=None)[source]¶
Get decoded expression.
- Parameters:
adata (
AnnData|None) – AnnData object with equivalent structure to initial AnnData. If None, defaults to the AnnData object used to initialize the model.indices (
Sequence[int] |None) – Indices of cells in adata to use. If None, all cells are used.batch_size (
int|None) – Minibatch size for data loading into model. Defaults to scvi.settings.batch_size.
- Return type:
- Returns:
Decoded expression for each cell
Examples
>>> import pertpy as pt >>> data = pt.dt.kang_2018() >>> pt.tl.SCGEN.setup_anndata(data, batch_key="label", labels_key="cell_type") >>> model = pt.tl.SCGEN(data) >>> model.train(max_epochs=10, batch_size=64, early_stopping=True, early_stopping_patience=5) >>> decoded_X = model.get_decoded_expression()
get_from_registry¶
- SCGEN.get_from_registry(adata, registry_key)¶
Returns the object in AnnData associated with the key in the data registry.
AnnData object should be registered with the model prior to calling this function via the
self._validate_anndatamethod.
get_latent_representation¶
- SCGEN.get_latent_representation(adata=None, indices=None, give_mean=True, n_samples=1, batch_size=None)[source]¶
Return the latent representation for each cell.
- Parameters:
adata (
AnnData|None) – AnnData object with equivalent structure to initial AnnData. If None, defaults to the AnnData object used to initialize the model.indices (
Sequence[int] |None) – Indices of cells in adata to use. If None, all cells are used.batch_size (
int|None) – Minibatch size for data loading into model. Defaults to scvi.settings.batch_size.
- Return type:
- Returns:
Low-dimensional representation for each cell
Examples
>>> import pertpy as pt >>> data = pt.dt.kang_2018() >>> pt.tl.SCGEN.setup_anndata(data, batch_key="label", labels_key="cell_type") >>> model = pt.tl.SCGEN(data) >>> model.train(max_epochs=10, batch_size=64, early_stopping=True, early_stopping_patience=5) >>> latent_X = model.get_latent_representation()
load¶
- classmethod SCGEN.load(dir_path, adata=None, accelerator='auto', device='auto', prefix=None, backup_url=None)¶
Instantiate a model from the saved output.
- Parameters:
dir_path (
str) – Path to saved outputs.adata (
Union[AnnData,MuData,None]) – AnnData organized in the same way as data used to train model. It is not necessary to run setup_anndata, as AnnData is validated against the saved scvi setup dictionary. If None, will check for and load anndata saved with the model.accelerator (
str) – Supports passing different accelerator types (“cpu”, “gpu”, “tpu”, “ipu”, “hpu”, “mps, “auto”) as well as custom accelerator instances.device (
int|str) – The device to use. Can be set to a non-negative index (int or str) or “auto” for automatic selection based on the chosen accelerator. If set to “auto” and accelerator is not determined to be “cpu”, then device will be set to the first available device.backup_url (
str|None) – URL to retrieve saved outputs from if not present on disk.
- Return type:
Model with loaded state dictionaries.
Examples
>>> model = ModelClass.load(save_path, adata) # use the name of the model class used to save >>> model.get_....
load_registry¶
plot_binary_classifier¶
- SCGEN.plot_binary_classifier(scgen, adata, delta, ctrl_key, stim_key, show=False, save=None, fontsize=14)[source]¶
Plots the dot product between delta and latent representation of a linear classifier.
Builds a linear classifier based on the dot product between the difference vector and the latent representation of each cell and plots the dot product results between delta and latent representation.
- Parameters:
scgen (
SCGEN) – ScGen object that was trained.adata (
AnnData|None) – AnnData object with equivalent structure to initial AnnData. If None, defaults to the AnnData object used to initialize the model. Must have been setup with batch_key and labels_key, corresponding to batch and cell type metadata, respectively.delta (
ndarray) – Difference between stimulated and control cells in latent spacectrl_key (
str) – Key for control part of the data found in condition_key.stim_key (
str) – Key for stimulated part of the data found in condition_key.path_to_save – Path to save the plot.
save (
str|bool|None) – Specify if the plot should be saved or not.fontsize (
float) – Set the font size of the plot.
- Return type:
plot_reg_mean_plot¶
- SCGEN.plot_reg_mean_plot(adata, condition_key, axis_keys, labels, save=None, gene_list=None, show=False, top_100_genes=None, verbose=False, legend=True, title=None, x_coeff=0.3, y_coeff=0.8, fontsize=14, **kwargs)[source]¶
Plots mean matching for a set of specified genes.
- Parameters:
adata – AnnData object with equivalent structure to initial AnnData. If None, defaults to the AnnData object used to initialize the model. Must have been setup with batch_key and labels_key, corresponding to batch and cell type metadata, respectively.
condition_key (
str) – The key for the conditionaxis_keys (
dict[str,str]) – Dictionary of adata.obs keys that are used by the axes of the plot. Has to be in the following form: {“x”: “Key for x-axis”, “y”: “Key for y-axis”}.labels (
dict[str,str]) – Dictionary of axes labels of the form {“x”: “x-axis-name”, “y”: “y-axis name”}.path_to_save – path to save the plot.
save (
str|bool|None) – Specify if the plot should be saved or not.show (
bool) – if True: will show to the plot after saving it.top_100_genes (
list[str]) – List of the top 100 differentially expressed genes. Specify if you want the top 100 DEGs to be assessed extra.verbose (
bool) – Specify if you want information to be printed while creating the plot, defaults to False.legend (
bool) – if True: plots a legend, defaults to True.title (
str) – Set if you want the plot to display a title.x_coeff (
float) – Offset to print the R^2 value in x-direction, defaults to 0.3.y_coeff (
float) – Offset to print the R^2 value in y-direction, defaults to 0.8.fontsize (
float) – Fontsize used for text in the plot, defaults to 14.**kwargs
- Return type:
Examples
>>> import pertpy as pt >>> data = pt.dt.kang_2018() >>> pt.tl.SCGEN.setup_anndata(data, batch_key="label", labels_key="cell_type") >>> scg = pt.tl.SCGEN(data) >>> scg.train(max_epochs=10, batch_size=64, early_stopping=True, early_stopping_patience=5) >>> pred, delta = scg.predict(ctrl_key='ctrl', stim_key='stim', celltype_to_predict='CD4 T cells') >>> pred.obs['label'] = 'pred' >>> eval_adata = data[data.obs['cell_type'] == 'CD4 T cells'].copy().concatenate(pred) >>> r2_value = scg.plot_reg_mean_plot(eval_adata, condition_key='label', axis_keys={"x": "pred", "y": "stim"}, labels={"x": "predicted", "y": "ground truth"}, save=False, show=True)
- Preview:
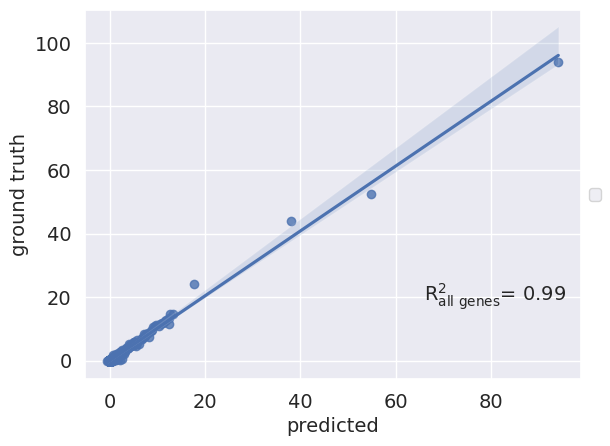
plot_reg_var_plot¶
- SCGEN.plot_reg_var_plot(adata, condition_key, axis_keys, labels, save=None, gene_list=None, top_100_genes=None, show=False, legend=True, title=None, verbose=False, x_coeff=0.3, y_coeff=0.8, fontsize=14, **kwargs)[source]¶
Plots variance matching for a set of specified genes.
- Parameters:
adata – AnnData object with equivalent structure to initial AnnData. If None, defaults to the AnnData object used to initialize the model. Must have been setup with batch_key and labels_key, corresponding to batch and cell type metadata, respectively.
condition_key (
str) – Key of the condition.axis_keys (
dict[str,str]) – Dictionary of adata.obs keys that are used by the axes of the plot. Has to be in the following form: {“x”: “Key for x-axis”, “y”: “Key for y-axis”}.labels (
dict[str,str]) – Dictionary of axes labels of the form {“x”: “x-axis-name”, “y”: “y-axis name”}.path_to_save – path to save the plot.
save (
str|bool|None) – Specify if the plot should be saved or not.show (
bool) – if True: will show to the plot after saving it.top_100_genes (
list[str]) – List of the top 100 differentially expressed genes. Specify if you want the top 100 DEGs to be assessed extra.legend (
bool) – if True: plots a legend, defaults to True.title (
str) – Set if you want the plot to display a title.verbose (
bool) – Specify if you want information to be printed while creating the plot, defaults to False.x_coeff (
float) – Offset to print the R^2 value in x-direction, defaults to 0.3.y_coeff (
float) – Offset to print the R^2 value in y-direction, defaults to 0.8.fontsize (
float) – Fontsize used for text in the plot, defaults to 14.
- Return type:
predict¶
- SCGEN.predict(ctrl_key=None, stim_key=None, adata_to_predict=None, celltype_to_predict=None, restrict_arithmetic_to='all')[source]¶
Predicts the cell type provided by the user in stimulated condition.
- Parameters:
ctrl_key – Key for control part of the data found in condition_key.
stim_key – Key for stimulated part of the data found in condition_key.
adata_to_predict – Adata for unperturbed cells you want to be predicted.
celltype_to_predict – The cell type you want to be predicted.
restrict_arithmetic_to – Dictionary of celltypes you want to be observed for prediction.
- Return type:
- Returns:
np nd-array of predicted cells in primary space.
- delta: float
Difference between stimulated and control cells in latent space
Examples
>>> import pertpy as pt >>> data = pt.dt.kang_2018() >>> pt.tl.SCGEN.setup_anndata(data, batch_key="label", labels_key="cell_type") >>> model = pt.tl.SCGEN(data) >>> model.train(max_epochs=10, batch_size=64, early_stopping=True, early_stopping_patience=5) >>> pred, delta = model.predict(ctrl_key="ctrl", stim_key="stim", celltype_to_predict="CD4 T cells")
register_manager¶
- classmethod SCGEN.register_manager(adata_manager)¶
Registers an
AnnDataManagerinstance with this model class.Stores the
AnnDataManagerreference in a class-specific manager store. Intended for use in thesetup_anndata()class method followed up by retrieval of theAnnDataManagervia the_get_most_recent_anndata_manager()method in the model init method.Notes
Subsequent calls to this method with an
AnnDataManagerinstance referring to the same underlying AnnData object will overwrite the reference to previousAnnDataManager.
save¶
- SCGEN.save(dir_path, prefix=None, overwrite=False, save_anndata=False, save_kwargs=None, **anndata_write_kwargs)¶
Save the state of the model.
Neither the trainer optimizer state nor the trainer history are saved. Model files are not expected to be reproducibly saved and loaded across versions until we reach version 1.0.
- Parameters:
dir_path (
str) – Path to a directory.prefix (
str|None) – Prefix to prepend to saved file names.overwrite (
bool) – Overwrite existing data or not. If False and directory already exists at dir_path, error will be raised.save_anndata (
bool) – If True, also saves the anndatasave_kwargs (
dict|None) – Keyword arguments passed intosave().anndata_write_kwargs – Kwargs for
write()
setup_anndata¶
- classmethod SCGEN.setup_anndata(adata, batch_key=None, labels_key=None, **kwargs)[source]¶
Sets up the
AnnDataobject for this model.A mapping will be created between data fields used by this model to their respective locations in adata. None of the data in adata are modified. Only adds fields to adata.
scGen expects the expression data to come from adata.X
- batch_key
key in adata.obs for batch information. Categories will automatically be converted into integer categories and saved to adata.obs[‘_scvi_batch’]. If None, assigns the same batch to all the data.
- labels_key
key in adata.obs for label information. Categories will automatically be converted into integer categories and saved to adata.obs[‘_scvi_labels’]. If None, assigns the same label to all the data.
Examples
>>> import pertpy as pt >>> data = pt.dt.kang_2018() >>> pt.tl.SCGEN.setup_anndata(data, batch_key="label", labels_key="cell_type")
to_device¶
- SCGEN.to_device(device)[source]¶
Move model to device.
- Parameters:
device – Device to move model to. Options: ‘cpu’ for CPU, integer GPU index (eg. 0), or ‘cuda:X’ where X is the GPU index (eg. ‘cuda:0’). See torch.device for more info.
Examples
>>> adata = scvi.data.synthetic_iid() >>> model = scvi.model.SCVI(adata) >>> model.to_device('cpu') # moves model to CPU >>> model.to_device('cuda:0') # moves model to GPU 0 >>> model.to_device(0) # also moves model to GPU 0
train¶
- SCGEN.train(max_epochs=None, accelerator='auto', devices='auto', train_size=0.9, validation_size=None, shuffle_set_split=True, batch_size=128, datasplitter_kwargs=None, plan_kwargs=None, **trainer_kwargs)¶
Train the model.
- Parameters:
max_epochs (
int|None) – Number of passes through the dataset. If None, defaults to np.min([round((20000 / n_cells) * 400), 400])accelerator (
str) – Supports passing different accelerator types (“cpu”, “gpu”, “tpu”, “ipu”, “hpu”, “mps, “auto”) as well as custom accelerator instances.devices (
int|list[int] |str) – The devices to use. Can be set to a non-negative index (int or str), a sequence of device indices (list or comma-separated str), the value -1 to indicate all available devices, or “auto” for automatic selection based on the chosen accelerator. If set to “auto” and accelerator is not determined to be “cpu”, then devices will be set to the first available device.train_size (
float) – Size of training set in the range [0.0, 1.0].validation_size (
float|None) – Size of the test set. If None, defaults to 1 - train_size. If train_size + validation_size < 1, the remaining cells belong to a test set.shuffle_set_split (
bool) – Whether to shuffle indices before splitting. If False, the val, train, and test set are split in the sequential order of the data according to validation_size and train_size percentages.batch_size (
int) – Minibatch size to use during training.lr – Learning rate to use during training.
datasplitter_kwargs (
dict|None) – Additional keyword arguments passed intoDataSplitter.plan_kwargs (
dict|None) – Keyword args forJaxTrainingPlan. Keyword arguments passed to train() will overwrite values present in plan_kwargs, when appropriate.**trainer_kwargs – Other keyword args for
Trainer.
view_anndata_setup¶
- SCGEN.view_anndata_setup(adata=None, hide_state_registries=False)¶
Print summary of the setup for the initial AnnData or a given AnnData object.