Note
This page was generated from sccoda.ipynb. Some tutorial content may look better in light mode.
scCODA - Compositional analysis of labeled single-cell data¶
This notebook serves as a tutorial for using the scCODA model (Büttner, Ostner et al., 2021) to analyze changes in cell composition data. Inspired by methods for compositional analysis of microbiome data, scCODA proposes a Bayesian approach to address the low replicate issue as commonly encountered in single-cell analysis. It models cell-type counts using a hierarchical Dirichlet-Multinomial model, which accounts for uncertainty in cell-type proportions and the negative correlative bias via joint modeling of all measured cell-type proportions. To ensure a uniquely identifiable solution and easy interpretability, the reference in scCODA is chosen to be a specific cell type. Hence, any detected compositional changes by scCODA always have to be viewed in relation to the selected reference.
Setup¶
[1]:
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
import mudata as mu
import pertpy as pt
Installed version 0.4.0 of pertpy is newer than the latest release 0.3.0! You are running a nightly version and features may break!
Data preparation¶
The data we use in the following example comes from Haber et al., 2017. It contains samples from the small intestinal epithelium of mice with different conditions. We first load the raw cell-level data. The dataset contains gene expressions of 9842 cells. They are annotated with their sample identifier (batch), the condition of the subjects and the type of each cell (cell_label).
[2]:
haber_cells = pt.dt.haber_2017_regions()
haber_cells.obs
[2]:
| batch | barcode | condition | cell_label | |
|---|---|---|---|---|
| index | ||||
| B1_AAACATACCACAAC_Control_Enterocyte.Progenitor | B1 | AAACATACCACAAC | Control | Enterocyte.Progenitor |
| B1_AAACGCACGAGGAC_Control_Stem | B1 | AAACGCACGAGGAC | Control | Stem |
| B1_AAACGCACTAGCCA_Control_Stem | B1 | AAACGCACTAGCCA | Control | Stem |
| B1_AAACGCACTGTCCC_Control_Stem | B1 | AAACGCACTGTCCC | Control | Stem |
| B1_AAACTTGACCACCT_Control_Enterocyte.Progenitor | B1 | AAACTTGACCACCT | Control | Enterocyte.Progenitor |
| ... | ... | ... | ... | ... |
| B10_TTTCACGACAAGCT_Salmonella_TA | B10 | TTTCACGACAAGCT | Salmonella | TA |
| B10_TTTCAGTGAGGCGA_Salmonella_Enterocyte | B10 | TTTCAGTGAGGCGA | Salmonella | Enterocyte |
| B10_TTTCAGTGCGACAT_Salmonella_Stem | B10 | TTTCAGTGCGACAT | Salmonella | Stem |
| B10_TTTCAGTGTGACCA_Salmonella_Endocrine | B10 | TTTCAGTGTGACCA | Salmonella | Endocrine |
| B10_TTTCAGTGTTCTCA_Salmonella_Enterocyte.Progenitor | B10 | TTTCAGTGTTCTCA | Salmonella | Enterocyte.Progenitor |
9842 rows × 4 columns
We now create an instance of a scCODA model and use it to aggregate the (cells x genes) data to a (samples x cell types) format by counting the cells for each category. The resulting MuData object sccoda_data includes both data aggregation levels as modalities rna and coda.
Looking at the data, we see that we have 4 control samples, and 3 conditions with 2 samples each. The resulting object separates our data components: Cell counts are stored in data.X, covariates in data.obs.
Further, it must contain one column or a set of columns (e.g. subject id, treatment, disease status) that uniquely identify each (statistical) sample. Further covariates (e.g. subject age) can either be specified via addidional column names in adata.obs, a key in adata.uns, or as a separate DataFrame.
[3]:
sccoda_model = pt.tl.Sccoda()
sccoda_data = sccoda_model.load(
haber_cells,
type="cell_level",
generate_sample_level=True,
cell_type_identifier="cell_label",
sample_identifier="batch",
covariate_obs=["condition"],
)
print(sccoda_data)
print(sccoda_data["coda"].X)
print(sccoda_data["coda"].obs)
Installed version 0.4.0 of pertpy is newer than the latest release 0.3.0! You are running a nightly version and features may break!
MuData object with n_obs × n_vars = 9852 × 15223
2 modalities
rna: 9842 x 15215
obs: 'batch', 'barcode', 'condition', 'cell_label', 'scCODA_sample_id'
coda: 10 x 8
obs: 'condition', 'batch'
var: 'n_cells'
[[ 36. 59. 136. 36. 239. 125. 191. 18.]
[ 32. 373. 116. 67. 117. 65. 168. 12.]
[ 5. 46. 23. 20. 50. 11. 40. 5.]
[ 45. 98. 188. 124. 250. 155. 365. 33.]
[ 26. 221. 198. 36. 131. 130. 196. 4.]
[ 52. 75. 347. 66. 323. 263. 313. 51.]
[ 65. 126. 115. 33. 65. 39. 129. 59.]
[ 42. 71. 203. 147. 271. 109. 180. 146.]
[ 40. 57. 383. 170. 321. 244. 256. 71.]
[ 37. 332. 113. 59. 90. 47. 132. 10.]]
condition batch
scCODA_sample_id
B1 Control B1
B10 Salmonella B10
B2 Control B2
B3 Control B3
B4 Control B4
B5 Hpoly.Day3 B5
B6 Hpoly.Day3 B6
B7 Hpoly.Day10 B7
B8 Hpoly.Day10 B8
B9 Salmonella B9
Note:
The term “samples” in scCODA refers to its statistical meaning. Each sample should contain all cells that were processed under the same conditions (e.g. same subject, treatment, disease status, …), which will be combined into one row of the aggregated data set. When using load, the key(s) to uniquely identify the samples must be specified in sample_identifier. Additional covariates that do not further distinguish the samples but are needed to create the model formula can be passed via
column names in adata.obs (covariate_obs), a key in adata.uns (covariate_uns), or as a separate DataFrame (covariate_df). When creating the aggregated dataset, scCODA will check the uniqueness of each additional covariate. Only if all cells in each sample have the same covariate value, it is possible to assign a value to the whole sample. If this is not the case, scCODA will skip the covariate with the message Covariate <xyz> has non-unique values! Skipping....
In our example, the column batch uniquely identifies each of the 10 samples, and we can pass condition as an additional covariate. To showcase the need for a list of sample_identifiers, consider the example where we obtained cells from three different subjects, which were split and then treated in three different ways:
cell |
subject |
treatment |
age |
|---|---|---|---|
cell1 |
S1 |
T1 |
35 |
cell2 |
S1 |
T3 |
35 |
cell3 |
S3 |
T2 |
42 |
cell4 |
S2 |
T1 |
50 |
cell5 |
S1 |
T1 |
35 |
In the aggregated data, we want nine samples (all combinations of subject and treatment). To achieve this, we need to pass the list ["subject", "treatment"] as the sample_identifier. The age column is unique for each subject (and therefore for each subject-treatment combination) and will thus be an additional covariate. The resulting aggregated obs DataFrame will be:
subject_treatment |
subject |
treatment |
age |
|---|---|---|---|
S1_T1 |
S1 |
T1 |
35 |
S1_T2 |
S1 |
T2 |
35 |
S1_T3 |
S1 |
T3 |
35 |
S2_T1 |
S2 |
T1 |
42 |
S2_T2 |
S2 |
T2 |
42 |
S2_T3 |
S2 |
T3 |
42 |
S3_T1 |
S3 |
T1 |
50 |
S3_T2 |
S3 |
T2 |
50 |
S3_T3 |
S3 |
T3 |
50 |
For our first example, we want to look at how the Salmonella infection influences the cell composition. Therefore, we create a subset of our compositional data that only contains the healthy and Salmonella-infected samples as a new data modality.
[4]:
# Select control and salmonella data
sccoda_data.mod["coda_salm"] = sccoda_data["coda"][
sccoda_data["coda"].obs["condition"].isin(["Control", "Salmonella"])
].copy()
print(sccoda_data["coda_salm"])
AnnData object with n_obs × n_vars = 6 × 8
obs: 'condition', 'batch'
var: 'n_cells'
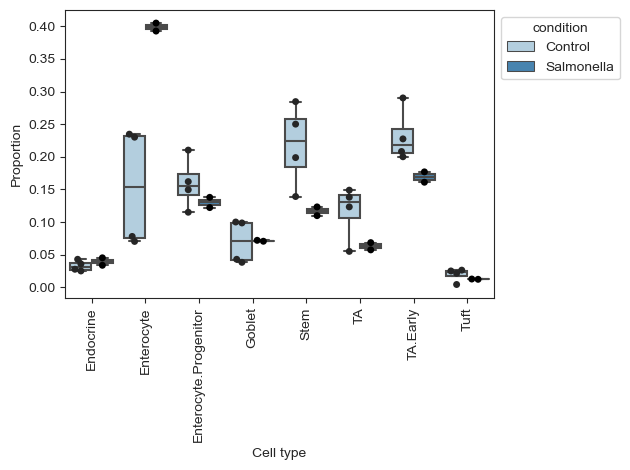
Plotting the data, we can see that there is a large increase of Enterocytes in the infected sampes, while most other cell types slightly decrease. Since scRNA-seq experiments are limited in the number of cells per sample, the count data is compositional, which leads to negative correlations between the cell types. Thus, the slight decreases in many cell types might be fully caused by the increase in Enterocytes.
[5]:
sccoda_model.plot_boxplots(sccoda_data, modality_key="coda_salm", feature_name="condition", add_dots=True)
plt.show()
Model setup and inference¶
We can now run parameter inference for the scCODA model to determine which cell types are credibly changing under Salmonella infection. To do this, we first need to specify all the necessary information through sccoda_model.prepare. We need to set:
The
formulaparameter. It specifies how the covariates are used in the model. It can process R-style formulas via the patsy package, e.g.formula="Cov1 + Cov2 + Cov3". Here, we simply use the “Condition” covariate of our datasetThe
reference_cell_typeparameter is used to specify a cell type that is believed to be unchanged by the covariates informula. This is necessary, because compositional analysis must always be performed relative to a reference (See Büttner, Ostner et al., 2021 for a more thorough explanation). If no knowledge about such a cell type exists prior to the analysis, taking a cell type that has a nearly constant relative abundance over all samples is often a good choice. It is also possible to let scCODA find a suited reference cell type by usingreference_cell_type="automatic". Here, we take Goblet cells as the reference.
[6]:
sccoda_data = sccoda_model.prepare(
sccoda_data,
modality_key="coda_salm",
formula="condition",
reference_cell_type="Goblet",
)
sccoda_data["coda_salm"]
[6]:
AnnData object with n_obs × n_vars = 6 × 8
obs: 'condition', 'batch'
var: 'n_cells'
uns: 'scCODA_params'
obsm: 'covariate_matrix', 'sample_counts'
No-U-turn HMC sampling is then initiated by calling sccoda_model.run_nuts().
[7]:
# Run MCMC
sccoda_model.run_nuts(sccoda_data, modality_key="coda_salm")
sccoda_data["coda_salm"]
sample: 100%|██████████| 11000/11000 [00:36<00:00, 303.04it/s, 127 steps of size 2.53e-02. acc. prob=0.83]
[7]:
AnnData object with n_obs × n_vars = 6 × 8
obs: 'condition', 'batch'
var: 'n_cells'
uns: 'scCODA_params'
obsm: 'covariate_matrix', 'sample_counts'
varm: 'intercept_df', 'effect_df_condition[T.Salmonella]'
Result interpretation¶
Calling sccoda_model.summary(), we can see the most relevant information for further analysis:
[8]:
sccoda_model.summary(sccoda_data, modality_key="coda_salm")
Compositional Analysis summary ┌──────────────────────────────────────────────┬──────────────────────────────────────────────────────────────────┐ │ Name │ Value │ ├──────────────────────────────────────────────┼──────────────────────────────────────────────────────────────────┤ │ Data │ Data: 6 samples, 8 cell types │ │ Reference cell type │ Goblet │ │ Formula │ condition │ └──────────────────────────────────────────────┴──────────────────────────────────────────────────────────────────┘
┌─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┐ │ Intercepts │ ├─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┤ │ Final Parameter Expected Sample │ │ Cell Type │ │ Endocrine 1.110 34.745 │ │ Enterocyte 2.319 116.401 │ │ Enterocyte.Progenitor 2.509 140.758 │ │ Goblet 1.741 65.303 │ │ Stem 2.693 169.193 │ │ TA 2.100 93.507 │ │ TA.Early 2.848 197.560 │ │ Tuft 0.426 17.532 │ └─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
┌─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┐ │ Effects │ ├─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┤ │ Final Parameter Expected Sample log2-fold change │ │ Covariate Cell Type │ │ conditionT.Salmonella Endocrine 0.000 24.831 -0.485 │ │ Enterocyte 1.352 321.438 1.465 │ │ Enterocyte.Progenitor 0.000 100.596 -0.485 │ │ Goblet 0.000 46.670 -0.485 │ │ Stem 0.000 120.918 -0.485 │ │ TA 0.000 66.827 -0.485 │ │ TA.Early 0.000 141.190 -0.485 │ │ Tuft 0.000 12.530 -0.485 │ └─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
Model properties
First, the summary shows an overview over the model properties:
Number of samples/cell types
The reference cell type.
The formula used
The model has two types of parameters that are relevant for analysis - intercepts and effects. These can be interpreted like in a standard regression model: Intercepts show how the cell types are distributed without any active covariates, effects show how the covariates influence the cell types.
Intercepts
The first column of the intercept summary shows the parameters determined by the MCMC inference.
The “Expected sample” column gives some context to the numerical values. If we had a new sample (with no active covariates) with a total number of cells equal to the mean sampling depth of the dataset, then this distribution over the cell types would be most likely.
Effects
For the effect summary, the first column again shows the inferred parameters for all combinations of covariates and cell types. Most important is the distinctions between zero and non-zero entries A value of zero means that no statistically credible effect was detected. For a value other than zero, a credible change was detected. A positive sign indicates an increase, a negative sign a decrease in abundance.
Since the numerical values of the “Final Parameter” column are not straightforward to interpret, the “Expected sample” and “log2-fold change” columns give us an idea on the magnitude of the change. The expected sample is calculated for each covariate separately (covariate value = 1, all other covariates = 0), with the same method as for the intercepts. The log-fold change is then calculated between this expected sample and the expected sample with no active covariates from the intercept section. Since the data is compositional, cell types for which no credible change was detected, can still change in abundance as well, as soon as a credible effect is detected on another cell type due to the sum-to-one constraint. If there are no credible effects for a covariate, its expected sample will be identical to the intercept sample, therefore the log2-fold change is 0.
Interpretation
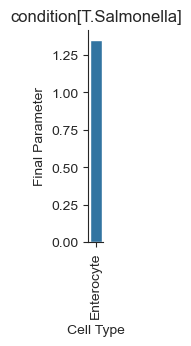
In the salmonella case, we see only a credible increase of Enterocytes, while all other cell types are unaffected by the disease. The log-fold change of Enterocytes between control and infected samples with the same total cell count lies at about 1.46.
We can also easily filter out all credible effects:
[9]:
sccoda_model.credible_effects(sccoda_data, modality_key="coda_salm")
[9]:
Covariate Cell Type
condition[T.Salmonella] Endocrine False
Enterocyte True
Enterocyte.Progenitor False
Goblet False
Stem False
TA False
TA.Early False
Tuft False
Name: Final Parameter, dtype: bool
We can also visualize the results as a barplot:
[10]:
sccoda_model.plot_effects_barplot(sccoda_data, modality_key="coda_salm", parameter="Final Parameter")
[10]:
<seaborn.axisgrid.FacetGrid at 0x7f96d999b580>
Adjusting the False discovery rate¶
scCODA selects credible effects based on their inclusion probability. The cutoff between credible and non-credible effects depends on the desired false discovery rate (FDR). A smaller FDR value will produce more conservative results, but might miss some effects, while a larger FDR value selects more effects at the cost of a larger number of false discoveries.
The desired FDR level can be easily set after inference via sim_results.set_fdr(). Per default, the value is 0.05, but we recommend to increase it up to 0.2 if no effects are found at a more conservative level.
In our example, setting a desired FDR of 0.4 reveals small effects on Endocrine and Tuft cells. Keep in mind that we chose this value only for instructive purposes, since there are no credible effects beside Enterocytes at lower FDR levels. In practice, expecting 40% of all credible effects to be false-positives is usually not recommended.
[11]:
sccoda_model.set_fdr(sccoda_data, modality_key="coda_salm", est_fdr=0.4)
sccoda_model.summary(sccoda_data, modality_key="coda_salm")
Compositional Analysis summary ┌──────────────────────────────────────────────┬──────────────────────────────────────────────────────────────────┐ │ Name │ Value │ ├──────────────────────────────────────────────┼──────────────────────────────────────────────────────────────────┤ │ Data │ Data: 6 samples, 8 cell types │ │ Reference cell type │ Goblet │ │ Formula │ condition │ └──────────────────────────────────────────────┴──────────────────────────────────────────────────────────────────┘
┌─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┐ │ Intercepts │ ├─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┤ │ Final Parameter Expected Sample │ │ Cell Type │ │ Endocrine 1.110 34.745 │ │ Enterocyte 2.319 116.401 │ │ Enterocyte.Progenitor 2.509 140.758 │ │ Goblet 1.741 65.303 │ │ Stem 2.693 169.193 │ │ TA 2.100 93.507 │ │ TA.Early 2.848 197.560 │ │ Tuft 0.426 17.532 │ └─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
┌─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┐ │ Effects │ ├─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┤ │ Final Parameter Expected Sample log2-fold change │ │ Covariate Cell Type │ │ conditionT.Salmonella Endocrine 0.282 32.626 -0.091 │ │ Enterocyte 1.352 318.408 1.452 │ │ Enterocyte.Progenitor 0.000 99.648 -0.498 │ │ Goblet 0.000 46.231 -0.498 │ │ Stem 0.000 119.778 -0.498 │ │ TA 0.000 66.197 -0.498 │ │ TA.Early 0.000 139.860 -0.498 │ │ Tuft -0.013 12.252 -0.517 │ └─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
Saving results¶
The MuData object containing all relevant information can be saved e.g. as a h5mu file. To access the saved data object, simply load it from disk and call the respective functions in an instance of a scCODA model
[12]:
# saving
path = "test"
sccoda_data.write_h5mu(path)
# loading
sccoda_data_2 = mu.read_h5mu(path)
sccoda_model.summary(sccoda_data_2, modality_key="coda_salm")
Compositional Analysis summary ┌──────────────────────────────────────────────┬──────────────────────────────────────────────────────────────────┐ │ Name │ Value │ ├──────────────────────────────────────────────┼──────────────────────────────────────────────────────────────────┤ │ Data │ Data: 6 samples, 8 cell types │ │ Reference cell type │ Goblet │ │ Formula │ condition │ └──────────────────────────────────────────────┴──────────────────────────────────────────────────────────────────┘
┌─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┐ │ Intercepts │ ├─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┤ │ Final Parameter Expected Sample │ │ Cell Type │ │ Endocrine 1.110 34.745 │ │ Enterocyte 2.319 116.401 │ │ Enterocyte.Progenitor 2.509 140.758 │ │ Goblet 1.741 65.303 │ │ Stem 2.693 169.193 │ │ TA 2.100 93.507 │ │ TA.Early 2.848 197.560 │ │ Tuft 0.426 17.532 │ └─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
┌─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┐ │ Effects │ ├─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┤ │ Final Parameter Expected Sample log2-fold change │ │ Covariate Cell Type │ │ conditionT.Salmonella Endocrine 0.282 32.626 -0.091 │ │ Enterocyte 1.352 318.408 1.452 │ │ Enterocyte.Progenitor 0.000 99.648 -0.498 │ │ Goblet 0.000 46.231 -0.498 │ │ Stem 0.000 119.778 -0.498 │ │ TA 0.000 66.197 -0.498 │ │ TA.Early 0.000 139.860 -0.498 │ │ Tuft -0.013 12.252 -0.517 │ └─────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘
[13]:
test_model = pt.tl.Sccoda()
[14]:
test_model.get_intercept_df(sccoda_data_2, modality_key="coda_salm")
[14]:
| Final Parameter | HDI 3% | HDI 97% | SD | Expected Sample | |
|---|---|---|---|---|---|
| Cell Type | |||||
| Endocrine | 1.110 | 0.448 | 1.808 | 0.366 | 34.745194 |
| Enterocyte | 2.319 | 1.734 | 2.913 | 0.315 | 116.401017 |
| Enterocyte.Progenitor | 2.509 | 1.898 | 3.093 | 0.316 | 140.757882 |
| Goblet | 1.741 | 1.126 | 2.330 | 0.320 | 65.303215 |
| Stem | 2.693 | 2.130 | 3.282 | 0.309 | 169.193202 |
| TA | 2.100 | 1.454 | 2.700 | 0.337 | 93.507465 |
| TA.Early | 2.848 | 2.269 | 3.427 | 0.308 | 197.559789 |
| Tuft | 0.426 | -0.274 | 1.162 | 0.391 | 17.532236 |
[15]:
test_model.get_effect_df(sccoda_data, modality_key="coda_salm")
[15]:
| Final Parameter | HDI 3% | HDI 97% | SD | Inclusion probability | Expected Sample | log2-fold change | ||
|---|---|---|---|---|---|---|---|---|
| Covariate | Cell Type | |||||||
| conditionT.Salmonella | Endocrine | 0.282468 | -0.476 | 1.097 | 0.319 | 0.4426 | 32.625962 | -0.090793 |
| Enterocyte | 1.351694 | 0.842 | 1.859 | 0.266 | 0.9996 | 318.408292 | 1.451774 | |
| Enterocyte.Progenitor | 0.000000 | -0.358 | 0.630 | 0.150 | 0.3100 | 99.647678 | -0.498308 | |
| Goblet | 0.000000 | 0.000 | 0.000 | 0.000 | 0.0000 | 46.230546 | -0.498308 | |
| Stem | 0.000000 | -0.805 | 0.193 | 0.217 | 0.3947 | 119.778085 | -0.498308 | |
| TA | 0.000000 | -0.843 | 0.382 | 0.238 | 0.3782 | 66.197370 | -0.498308 | |
| TA.Early | 0.000000 | -0.465 | 0.451 | 0.126 | 0.2806 | 139.859835 | -0.498308 | |
| Tuft | -0.012933 | -0.892 | 0.987 | 0.312 | 0.3994 | 12.252233 | -0.516965 |
[16]:
sccoda_data["coda_salm"].varm["intercept_df"]
[16]:
| Final Parameter | HDI 3% | HDI 97% | SD | Expected Sample | |
|---|---|---|---|---|---|
| Cell Type | |||||
| Endocrine | 1.110 | 0.448 | 1.808 | 0.366 | 34.745194 |
| Enterocyte | 2.319 | 1.734 | 2.913 | 0.315 | 116.401017 |
| Enterocyte.Progenitor | 2.509 | 1.898 | 3.093 | 0.316 | 140.757882 |
| Goblet | 1.741 | 1.126 | 2.330 | 0.320 | 65.303215 |
| Stem | 2.693 | 2.130 | 3.282 | 0.309 | 169.193202 |
| TA | 2.100 | 1.454 | 2.700 | 0.337 | 93.507465 |
| TA.Early | 2.848 | 2.269 | 3.427 | 0.308 | 197.559789 |
| Tuft | 0.426 | -0.274 | 1.162 | 0.391 | 17.532236 |
References¶
Büttner, M., Ostner, J., Müller, C.L. et al. scCODA is a Bayesian model for compositional single-cell data analysis. Nat Commun 12, 6876 (2021). https://doi.org/10.1038/s41467-021-27150-6
Haber, A., Biton, M., Rogel, N. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). https://doi.org/10.1038/nature24489